|
|
Genetika
→
Articles
ArticlesPossible Function of the ribT Gene of Bacillus subtilis: Theoretical Prediction, Cloning, and Expression.Possible Function of the ribT Gene of Bacillus subtilis: Theoretical Prediction, Cloning, and Expression.
A P Yakimov T A Seregina A A Kholodnyak R A Kreneva A S Mironov D A Perumov A L Timkovskii Acta naturae 07/2014; 6(3):106-9. Source: PubMed ABSTRACT The complete decipherment of the functions and interactions of the elements of the riboflavin biosynthesis operon (rib operon) of Bacillus subtilis are necessary for the development of superproducers of this important vitamin. The function of its terminal ribT gene has not been established to date. In this work, a search for homologs of the hypothetical amino acid sequence of the gene product through databases, as well as an analysis of the homolgs, was performed; the distribution of secondary structure elements was theoretically predicted; and the tertiary structure of the RibT protein was proposed. The ribT gene nucleotide sequence was amplified and cloned into the standard high-copy expression vector pET15b and then expressed after induction with IPTG in E. coli BL21 (DE3) strain cells containing the inducible phage T7 RNA polymerase gene. The ribT gene expression was confirmed by SDS-PAGE. The protein product of the expression was purified by affinity chromatography. Therefore, the real possibility of RibT protein production in quantities sufficient for further investigation of its structure and functional activity was demonstrated. 12.01.2015
Optimization of genetic constructs for high-level expression of the darbepoetin gene in mammalian cellsR. R. Shukurov, K. Yu. Kazachenko, D. G. Kozlov, A. A. Nurbakov, E. N. Sautkina, R. A. Khamitov, Yu. A. Seryogin, Applied Biochemistry and Microbiology (Impact Factor: 0.66). 12/2014; 50(9). DOI: 10.1134/S0003683814090051 ABSTRACT Genetic constructs were designed in order to optimize darbepoetin production in CHO cells. They are characterized by a higher level of structural optimization of the darbepoetin gene, a higher gene dose, and the selection of promoter elements that have never been used before for this purpose. A transient transfection of CHO cells by the obtained constructs was performed. It was shown that each of the variable factors in the constructs influenced darbepoetin gene expression. A construct containing a doubled dose of the darbepoetin synthetic gene with optimized codon composition under the control of the CMV-EF1α chimerical promoter was proved to be the most efficient. 12.01.2015
Riboswitches in regulation of Rho-dependent transcription termination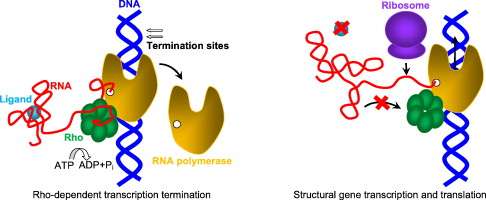
Sergey Proshkin, Alexander Mironov, Evgeny Nudler Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms (Impact Factor: 5.46). 01/2014; 27.04.2014
Comparison of different approaches to activate the glyoxylate bypass in Escherichia coli K-12 for succinate biosynthesis during dual-phase fermentation in minimal glucose media.Alexandra Yu Skorokhodova, Andrey Yu Gulevich, Anastasiya A Morzhakova, Rustem S Shakulov, Vladimir G Debabov ABSTRACT: Two different approaches to activate the glyoxylate bypass in model Escherichia coli K-12 strains for succinate biosynthesis during dual-phase fermentation in minimal glucose media were examined. Inactivation of IclR and FadR, the transcriptional regulators of the aceBAK operon, were insufficient for the involvement of the glyoxylate bypass in anaerobic succinate biosynthesis by strains grown aerobically under glucose-abundant conditions. In contrast, the strains that constitutively expressed the aceEF-lpdA operon coding for the pyruvate dehydrogenase complex could partially synthesise succinate anaerobically via the glyoxylate bypass, even in the presence of intact regulators. The results suggest that the intensive acetyl-CoA formation in the strains constitutively expressing pyruvate dehydrogenase matches the physiological conditions that favour the activation of the glyoxylate bypass. Biotechnology Letters 12/2012 03.09.2013
Microbial Producers of ButanolO. V. Berezinaa, N. V. Zakharovaa, C. V. Yarotskya, and V. V. Zverlovb a State Research Institute of Genetics and Selection of Industrial Microorganisms, Moscow, 117545 Russia b Technische Universität München, Department of Microbiology, Freising, 85350 Germany Abstract—This review is written due to an increased interest in the production of energy carriers and basic substrates of the chemical industry from renewable natural resources. In this review, the microbiological aspects of biobutanol production are reflected and the microbial producers of butanol (both natural, i.e., members of the Clostridium genus, and recombinant), obtained by genetic modification of Clostridia and other microorganisms, are characterised. Keywords: biofuel, butanol, renewable feedstock, clostridia, recombinant butanol producers, Clostridium acetobutylicum 25.08.2013
Bacterial Nitric Oxide Extends the Lifespan of C. elegansIvan Gusarov, Laurent Gautier, Olga Smolentseva, Ilya Shamovsky, Svetlana Eremina, Alexander Mironov, Evgeny Nudler Department of Biochemistry and Molecular Pharmacology, New York University School of Medicine, New York, NY 10016, USA. Cell (impact factor: 32.4). 02/2013; 152(4):818-30. DOI:10.1016/j.cell.2012.12.043 Source: PubMed ABSTRACT Nitric oxide (NO) is an important signaling molecule in multicellular organisms. Most animals produce NO from L-arginine via a family of dedicated enzymes known as NO synthases (NOSes). A rare exception is the roundworm Caenorhabditis elegans, which lacks its own NOS. However, in its natural environment, C. elegans feeds on Bacilli that possess functional NOS. Here, we demonstrate that bacterially derived NO enhances C. elegans longevity and stress resistance via a defined group of genes that function under the dual control of HSF-1 and DAF-16 transcription factors. Our work provides an example of interspecies signaling by a small molecule and illustrates the lifelong value of commensal bacteria to their host. 25.08.2013
Article: Semi-bleached paper and fermentation products from a larch biorefineryHanna Horhammer, Oksana Berezina, Eero Hiltunen, Tom Granstrom, Adriaan Van Heiningen ABSTRACT: This study was focused on the products from a larch biorefinery, specifically bleached paper and different fermentation products. Siberian larch (Larix sibirica Lebed.) wood chips were treated with water in a pre-extraction (PE) stage. The larch extract was removed by drainage and fermented into different products. Eight different bacteria strains were tested. The extracted wood chips were mildly washed before kraft pulping with polysulfide (PS) and anthraquinone (AQ). The PE-PSAQ pulps were bleached to about 80% brightness. Laboratory paper sheets were made and tested for different paper properties, and a conventional larch kraft pulp was also prepared as reference. The larch PE-PSAQ paper and the larch kraft paper had similar properties. The removal of a significant amount of hemicelluloses from the wood chips before pulping was not a detriment to the paper properties. Application: Combining papermaking pulp production with production of value-added products from the extracted hemicellulose stream resulted in a promising biorefinery concept. Tappi Journal 10/2012; 11(10). 25.08.2013
Methylation of the Promoter Region of the RASSF1A Candidate Tumor Suppressor Gene in Primary Epithelial TumorsV. I. Loginov, A. V. Malyukova, Y. A. Seryogin, D. S. Hodyrev, T. P. Kazubskaya, V. D. Ermilova, R. F. Garkavtseva, L. L. Kisselev, E. R. Zabarovsky, E. A. Braga State Research Center GosNIIgenetika; Russian Academy of Medical Sciences; Russian Academy of Sciences; Karolinska Institute Molecular Biology (impact factor: 0.66). 06/2004; 38(4):549-560. DOI:10.1023/B:MBIL.0000037007.71787.b9 ABSTRACT The methylation of the promoter CpG island of the RASSF1A tumor suppressor gene in primary tumors of 172 Muscovites with renal cell carcinoma (RCC), breast cancer (BC), or ovarian epithelial tumors (OET) was assayed by means of methylation-specific PCR (MSP) and PCR-based methylation-sensitive restriction enzyme analysis (MSRA). The MSP, MSRA, and previous bisulfite sequencing data correlated significantly with each other (P 10–6 for Spearman's rank correlation coefficients). By MSP and MSRA, the respective methylation frequencies of the RASSF1A promoter were 86% (25/29) and 94% (50/53) in RCC, 64% (18/28) and 78% (32/41) in BC, and 59% (17/29) and 73% (33/45) in OET. Methylation-sensitive restriction enzymes (HpaII, HhaI, Bsh1236I, AciI) increased the analysis sensitivity and made it possible to establish the methylation status for 18 CpG dinucleotides of the RASSF1A promoter region. With the MSRA data, the density of methylation of the CpG island was estimated at 72% in RCC, 63% in BC, and 58% in OET (the product of the number of CpG dinucleotides and the number of specimens with RASSF1A methylation was taken as 100%). Methylation of the RASSF1A promoter region was observed in 11–35% of the DNA specimens from the histologically normal tissue adjacent to the tumor but not in the peripheral blood DNA of 15 healthy subjects. The RASSF1A methylation frequency showed no significant correlation with the stage, grade, and metastatic potential of the tumor. On the other hand, epigenetic modification of RASSF1A was considerably more frequent than hemizygous or homozygous deletions from the RASSF1A region. These results testify that methylation of the RASSF1A promoter region takes place early in carcinogenesis and is a major mechanism inactivating RASSF1A in epithelial tumors. 21.03.2013
New Tumor Suppressor Genes in Hot Spots of Human Chromosome 3: New Methods of IdentificationE. A. Braga, V. I. Kashuba, A. V. Malyukova, V. I. Loginov, V. N. Senchenko, I. V. Bazov, L. L. Kisselev, E. R. Zabarovsky State Research Center GosNIIgenetika; Karolinska Institute; National Academy of Sciences of Ukraine; Russian Academy of Sciences Molecular Biology (impact factor: 0.66). 01/2003; 37(2):170-185. DOI:10.1023/A:1023381218481 ABSTRACT Studies of the recent decade, including sequencing of numerous human genome regions, allowed a great progress in detection of new tumor suppressor genes (TSG) and development of new means of their identification and analysis. Effective methods of genome scanning and TSG identification combine DNA array techniques and subtraction hybridization. Alternative ways take advantage of new extrachromosomal vector systems (pETE, pETR) and the functional gene inactivation test. A breakthrough was made in localizing new TSG on the human chromosome 3 short arm, which harbors tumor-suppressing regions and is often rearranged in various tumors and in early carcinogenesis. On 3p, only three putative TSG were known five years ago, and at least ten were identified by the end of 2002. The role of new TSG in carcinogenesis is commonly inferred from a decrease in their transcription in tumor cell lines or primary tumors and from their ability to suppress the tumor growth. Protein products of 3p TGS perform an important function, constraining cell malignization. Some of them are directly involved in regulating the cell cycle and inducing apoptosis (RASSFIA), others suppress angiogenesis (Sema3B) or metastasis (Hyal-1). Numerous attempts to find mutations in exons of silenced genes failed, and at least half of the new candidate genes (RASSFIA, CACNA2D2, BLU, HYAL1, SEMA3B, RAR-) proved to be inactivated by promoter methylation. 21.03.2013
Adaptation of acrylamide producer Rhodococcus rhodochrous M8 to change in ammonium concentration in mediumO B Astaurova, T E Leonova, I N Poliakova, I V Sineokaia, V K Gordeev, A S Ianenko State Research Institute of Genetics and Selection of Industrial Microorganisms, Moscow, Russia. Prikladnaia biokhimiia i mikrobiologiia 36(1):21-5. Source: PubMed ABSTRACT The mechanism of adaptation of the acrylamide producing strain Rhodococcus rhodochrous M8 to changes in ammonium concentrations in the medium was studied. An increase in the content of ammonium in the medium changed the activity of glutamine synthetase (GS) (EC 6.3.1.2) and glutamine dehydrogenase (GD) (EC 1.4.1.4), the enzymes of ammonium assimilation, as well as the activities of enzymes responsible for nitrile utilization: nitrile hydratase (EC 4.2.1.84) and amidase (EC 3.5.1.4). This also caused inhibition of activation of GS induced by phosphodiesterase (EC 3.1.4.1). Increases in the activities of nitrile hydratase and amidase and resistance of these enzymes to ammonium were observed in mutant of R. rhodichrous resistant to phosphotricine, an inhibitor of GS. An important role of GS in the mechanism of adaptation is suggested. 21.03.2013
Isolation and primary characterization of an amidase from Rhodococcus rhodochrousE K Kotlova, G G Chestukhina, O B Astaurova, T E Leonova, A S Yanenko, V G Debabov Institute of Genetics and Selection of Industrial Microorganisms, Moscow, 113545, Russia. . Biochemistry (Moscow) (impact factor: 1.06). 05/1999; 64(4):384-9. Source: PubMed ABSTRACT Amidase (EC 3.5.1.4) was purified to homogeneity from Rhodococcus rhodochrous M8 using isopropanol fractionation and exchange chromatography on Mono Q. The isolated amidase consists of four identical subunits with molecular weight 42+/-2 kD. The activity of the enzyme is maximal at 55-60 degrees C and within the pH range 5-8. The amidase from R. rhodochrous M8 is highly sensitive to such sulfhydryl reagents as Hg2+ and Cu2+. Chelators (EDTA and o-phenanthroline) and serine proteinase inhibitors (PMSF and DIFP) did not inhibit the activity of the enzyme. The enzyme exhibits hydrolytic and acyl transferase activity and does not possess urease activity. Aliphatic amides (acetamide and propionamide) were the best substrates for the amidase from R. rhodochrous M8, whereas bulky aromatic amides were poor substrates of this enzyme. The properties of the isolated enzyme are similar to those found in the corresponding amidase from Arthrobacter sp. J-1 and an amidase with wide substrate specificity from Brevibacterium sp. R312. 13.03.2013
The riboswitch-mediated control of sulfur metabolism in bacteriaVitaly Epshtein, Alexander S Mironov, Evgeny Nudler Department of Biochemistry, New York University Medical Center, New York, NY 10016, USA. Proceedings of the National Academy of Sciences (impact factor: 9.68). 05/2003; 100(9):5052-6. DOI:10.1073/pnas.0531307100 Source: PubMed ABSTRACT Many operons in Gram-positive bacteria that are involved in methionine (Met) and cysteine (Cys) biosynthesis possess an evolutionarily conserved regulatory leader sequence (S-box) that positively controls these genes in response to methionine starvation. Here, we demonstrate that a feed-back regulation mechanism utilizes S-adenosyl-methionine as an effector. S-adenosyl-methionine directly and specifically binds to the nascent S-box RNA, causing an intrinsic terminator to form and interrupt transcription prematurely. The S-box leader RNA thus expands the family of newly discovered riboswitches, i.e., natural regulatory RNA aptamers that seem to sense small molecules ranging from amino acid derivatives to vitamins. 11.03.2013
Riboswitch control of Rho-dependent transcription terminationKerry Hollands, Sergey Proshkin, Svetlana Sklyarova, Vitaly Epshtein, Alexander Mironov, Evgeny Nudler, Eduardo A Groisman Section of Microbial Pathogenesis, Yale School of Medicine, New Haven, CT 06536, USA. Proceedings of the National Academy of Sciences (impact factor: 9.68). 03/2012; 109(14):5376-81. DOI:10.1073/pnas.1112211109 Source: PubMed ABSTRACT Riboswitches are RNA sensors that regulate gene expression upon binding specific metabolites or ions. Bacterial riboswitches control gene expression primarily by promoting intrinsic transcription termination or by inhibiting translation initiation. We now report a third general mechanism of riboswitch action: governing the ability of the RNA-dependent helicase Rho to terminate transcription. We establish that Rho promotes transcription termination in the Mg(2+)-sensing mgtA riboswitch from Salmonella enterica serovar Typhimurium and the flavin mononucleotide-sensing ribB riboswitch from Escherichia coli when the corresponding riboswitch ligands are present. The Rho-specific inhibitor bicyclomycin enabled transcription of the coding regions at these two loci in bacteria experiencing repressing concentrations of the riboswitch ligands in vivo. A mutation in the mgtA leader that favors the "high Mg(2+)" conformation of the riboswitch promoted Rho-dependent transcription termination in vivo and in vitro and enhanced the ability of the RNA to stimulate Rho's ATPase activity in vitro. These effects were overcome by mutations in a C-rich region of the mRNA that is alternately folded at high and low Mg(2+), suggesting a role for this region in regulating the activity of Rho. Our results reveal a potentially widespread mode of gene regulation whereby riboswitches dictate whether a protein effector can interact with the transcription machinery to prematurely terminate transcription. 11.03.2013
Construction of a butyrate-producing E. coli strain without the use of heterologous genesT. A. Seregina, R. S. Shakulov, V. G. Debabov, A. S. Mironov ABSTRACT: Multistage construction of an E. coli strain containing no foreign genes which is capable of producing butyrate has been carried out. At the first stage, deletions in gene fadR encoding a protein repressor of an operon for fatty acid degradation and gene aceF responsible for the synthesis of pyruvate dehydrogenase were introduced in the strain MG1655 genome. Then, a mutant obtained from the above strain by induced mutagenesis and capable of growth on ethanol as a sole carbon source under aerobic conditions was selected. It was shown that growth of the mutant on ethanol is provided by two mutations. One of them (a substitution: 257G → A) is located in the regulatory region of gene adhE that controls the synthesis of alcohol-dehydrogenase; the other, containing a substitution Glu568 → Lys, affects the structural portion of the gene. As a result of the consequent mutagenesis of the obtained strain and selection on indicating media, variants capable of growing on butyrate and butanol as sole carbon sources and putatively bearing mutations in gene atoC (encoding transcriptional activator of atoDAB operon) were selected. At the last stage of the work, gene atoB, encoding the synthesis of the thiolase II enzyme, was placed under the control of a constitutive promoter P tet , and the functional allele of gene aceF was introduced. The resulting E. coli strain (ΔfadR, adhE, atoC, P tet -atoB) accumulates 800 mg/l of butyrate upon growth on glucose-containing medium under semi-anaerobic (oxygen limited) conditions. Introduction of an additional deletion in gene ldhA encoding lactate dehydrogenase in the strain genome leads to a further growth of a butyrate production up to 1.3 g/l. Key wordsgenetic construction-fatty acid β-oxidation-butyrate synthesis- E. coli Applied Biochemistry and Microbiology 04/2012; 46(8):745-754. 07.03.2013
Multifunctional regulatory mutation in Bacillus subtilis flavinogenesis systemR. A. Kreneva, D. V. Karelov, N. V. Korolkova, A. S. Mironov, D. A. Perumov ABSTRACT: Among Bacillus subtilis riboflavin-resistant mutants we identified one, which differed from other regulatory mutants by overproduction of riboflavin and simultaneous upregulation of the ribC gene encoding flavokinase/FAD-synthase. Genetic and biochemical analysis showed that the ribU1 mutation determines a trans-acting factor that simultaneously regulates activity of riboflavin and truB-ribC-rpsO operons. Regulatory activity of the ribU1 mutation comprises about 10% of Rfn element activity on interaction with flavins. The ribU1 mutation can be presumably ascribed to a gene of the transcriptional regulators family. Russian Journal of Genetics 04/2012; 45(10):1256-1259. 07.03.2013
Cloning and expression of bacteriophage FMV lysocyme gene in cells of yeasts Saccharomyces cerevisiae and Pichia pastorisD. G. Kozlov, S. E. Cheperegin, A. V. Chestkov, V. N. Krylov, Yu. D. Tsygankov ABSTRACT: Cloning, sequencing, and expression of the gene for soluble lysozyme of bacteriophage FMV from Gram-negative Pseudomonas aeruginosa bacteria were conducted in yeast cells. Comparable efficiency of two lysozyme expression variants (as intracellular or secreted proteins) was estimated in cells of Saccharomyces cerevisiae and Pichia pastoris. Under laboratory conditions, yeast S. cerevisiae proved to be more effective producer of phage lysozyme than P. pastoris, the yield of the enzyme in the secreted form being significantly higher than that produced in the intracellular form. Russian Journal of Genetics 04/2012; 46(3):300-307. 07.03.2013
Novel method for screening Saccharomyces cerevisiae mutants with increased sulfur-containing compounds: Color-based selection of ade1 or ade2 mutants.Marina G Tarutina, Tatiana A Dutova, Inna E Yezhova, Hiroaki Nishiuchi, Sergey P Sineoky [hide abstract] ABSTRACT: We identified Saccharomyces cerevisiae mutants with 100% higher intracellular glutathione using 1-methyl-3-nitro-1-nitrosoguanidine mutagenesis. This method employs visual selection of the most pigmented colonies among met30 strains carrying ade1 and ade2 mutations. Since the method does not involve genetic engineering, the mutants are suitable for use in the food industry. Journal of Bioscience and Bioengineering 08/2012; · 1.79 Impact Factor 21.08.2012
Cloning and characterisation of a large metagenomic DNA fragment containing glycosyl-hydrolase genesE. N. Shedova, O. V. Berezina, N. A. Lunina, V. V. Zverlov, W. H. Schwarz, G. A. Velikodvorskaya ABSTRACT: The problem of searching for and characterizing enzymes produced by uncultured microorganisms is presently settled by creating metagenomic libraries. A 6000-clone library with the average size of inserts of about 15 kb has been constructed based on total DNA isolated from cow rumen microorganisms. As a result of library screening on plates with different substrates, a clone was selected that efficiently hydrolysed lichenan and carboxymethylcellulose. The clone contained the recombinant plasmid pBlue-13 bearing a 12071 bp-long metagenomic fragment carrying ten open reading frames, two of which were identified as glycosyl hydrolase genes. No homology of the metagenomic DNA with any sequences known microorganism genomes was revealed. The amino acid sequence deduced from frame 4 was denoted of Xyl3A, and bears resemblance with β-xylosidases of glycosyl hydrolase family 3. Frame 6 encodes polypeptide Cel5A homologous to cellulases of glycosyl hydorlase family 5. The amino acid sequences deducted from on seven out of ten open reading frames were homologous to proteins of microorganisms belonging to the Bacteroides sp. family and the bacteria inhabiting mammalian intestines. Molecular Genetics Microbiology and Virology 05/2012; 24(1):12-16. 07.05.2012
Biodegradable matrices from regenerated silk of Bombix moriI I Agapov, M M Moisenovich, T V Vasilyeva, O L Pustovalova, A S Kon'kov, A Yu Arkhipova, O S Sokolova, V G Bogush, V I Sevastianov, V G Debabov, M P Kirpichnikov Doklady Biochemistry and Biophysics 04/2012; 433:201-4. · 0.33 Impact Factor 18.04.2012
Lux-biosensors for detection of SOS-response, heat shock, and oxidative stressV. Yu. Kotova, I. V. Manukhov, G. B. Zavilgelskii ABSTRACT: We have constructed hybrid plasmids pColD and pIbpA in which transcription of reporter genes luxCDABE from Photohabdus luminescens occurred from induced promoters: SOS-promoter of gene cdaA (pColD-CA23) and heat shock gene ibpA (E. coli MG1655 genome), respectively. Main parameters were measured: threshold concentration of an inducer and maximal response and minimal time of response for E. coli lux-biosensors bearing hybrid plasmids pColD and pRecA (SOS-response); pIbpA and pGrpe (heat shock) and pKatG and pSoxS (oxidative stress). Mitomicyn C, cis-diaminedichloroplatinum (SOS-response), ethanol and pentachlorophenol (heat shock), and hydrogen peroxide and methyl viologen (oxidative stress) were used as luminescence inducers. It was shown that the SOS-biosensor pColD is superior in main characteristics to the earlier constructed biosensor pRecA, and the heat biosensor pIbpA is more efficient than the earlier constructed pGrpE. Lux-biosensors (pKatG) and (pSoxS) are high sensitive to hydrogen peroxide and methyl viologen, respectively. Key wordsbioluminescence-biosensor-induced promoter-Pcda-PibpA-PgrpE-PkatG-PrecA-PsoxS Applied Biochemistry and Microbiology 04/2012; 46(8):781-788. 07.04.2012
Lux-biosensors for detection of SOS-response, heat shock, and oxidative stressV. Yu. Kotova, I. V. Manukhov, G. B. Zavilgelskii ABSTRACT: We have constructed hybrid plasmids pColD and pIbpA in which transcription of reporter genes luxCDABE from Photohabdus luminescens occurred from induced promoters: SOS-promoter of gene cdaA (pColD-CA23) and heat shock gene ibpA (E. coli MG1655 genome), respectively. Main parameters were measured: threshold concentration of an inducer and maximal response and minimal time of response for E. coli lux-biosensors bearing hybrid plasmids pColD and pRecA (SOS-response); pIbpA and pGrpe (heat shock) and pKatG and pSoxS (oxidative stress). Mitomicyn C, cis-diaminedichloroplatinum (SOS-response), ethanol and pentachlorophenol (heat shock), and hydrogen peroxide and methyl viologen (oxidative stress) were used as luminescence inducers. It was shown that the SOS-biosensor pColD is superior in main characteristics to the earlier constructed biosensor pRecA, and the heat biosensor pIbpA is more efficient than the earlier constructed pGrpE. Lux-biosensors (pKatG) and (pSoxS) are high sensitive to hydrogen peroxide and methyl viologen, respectively. Key wordsbioluminescence-biosensor-induced promoter-Pcda-PibpA-PgrpE-PkatG-PrecA-PsoxS Applied Biochemistry and Microbiology 04/2012; 46(8):781-788. 07.04.2012
Three-dimensional scaffold made from recombinant spider silk protein for tissue engineeringI. I. Agapov, O. L. Pustovalova, M. M. Moisenovich, V. G. Bogush, O. S. Sokolova, V. I. Sevastyanov, V. G. Debabov, M. P. Kirpichnikov Doklady Biochemistry and Biophysics 04/2012; 426(1):127-130. 07.04.2012
Adaptation of Rhodococcus rhodochrous M8, a producer of acrylamide, to changes in ammonium concentration in the growth mediumO. B. Astaurova, T. E. Leonova, I. N. Polyakova, I. V. Sineokaya, V. K. Gordeev, A. S. Yanenko ABSTRACT: The mechanism of adaptation of the acrylamide producing strainRhodococcus rhodochrous M8 to changes in ammonium concentrations in the medium was studied. An increase in the content of ammonium in the medium changed the activity of glutamine synthetase (GS) (EC 6.3.1.2) and glutamine dehydrogenase (GD) (EC 1.4.1.4), the enzymes of ammonium assimilation, as well as the activities of enzymes responsible for nitrile utilization: nitrile hydratase (EC 4.2.1.84) and amidase (EC 3.5.1.4). This also inhibited the activation of GS induced by phosphodiesterase (EC 3.1.4.1 ). Increases in the activities of nitrile hydratase and amidase and resistance of these enzymes to ammonium were observed in mutant ofR. rhodichrous resistant to phosphotricine, an inhibitor of GS. An important role of GS in the mechanism of adaptation is suggested. Applied Biochemistry and Microbiology 04/2012; 36(1):15-18. 07.04.2012
Human chromosome 3P regions of putative tumor-suppressor genes in renal, breast, and ovarian carcinomasV. I. Loginov, I. V. Bazov, D. S. Khodyrev, I. V. Pronina, T. P. Kazubskaya, V. D. Ermilova, R. F. Garkavtseva, E. R. Zabarovsky, E. A. Braga ABSTRACT: Allelic imbalances (AI) of polymorphic markers at the short arm of chromosome 3 (3p) were mapped using DNA samples of renal cell carcinoma (RCC, 80 cases), breast carcinoma (BC, 95 cases), and epithelial ovarian cancer (EOC, 50 cases) at the same dense panel of markers (up to 24 loci). Six regions with the increased AI frequency (versus the average values determined for all the analyzed 3p markers) at RCC, BC or EOC were found in 3p chromosome. Four 3p regions presumably contain tumor-suppressor genes (TSG) involved in the epithelial tumors of various types. Region between D3S2409 and D3S3667 markers in the 3p21.31 region was identified in this study for the first time. The AI peak in D3S2409-D3S3667 region was statistically significant (P < 0.001, according to Fisher) when representative sample set of 95 BC patients was analyzed. The data on increased frequency of polymorphic marker allele amplification suggest that the D3S2409-D3S3667 region contains both putative TSG and protooncogenes. Russian Journal of Genetics 04/2012; 44(2):209-214. 07.04.2012
Titanium dioxide (TiO2) nanoparticles induce bacterial stress response detectable by specific lux biosensorsG. B. Zavilgelsky, V. Yu. Kotova, I. V. Manukhov ABSTRACT: To investigate the mechanism of the bactericidal effect of TiO2 nanoparticles, we used induced lux biosensors on the basis of the Escherichia coli MG1655 strain containing pColD-lux, pKatG-lux, and pSoxS-lux plasmids. It was shown that the UV-light (λ = 360–390 nm) treatment of a suspension of biosensor cells and TiO2 nanoparticles induces H2O2 formation in bacteria, which induces the expression of the luxCDABE reporter genes, leading to bioluminescence intensification. It is proposed to use the specifically induced lux biosensors for the detection of photoactive dioxide metal nanoparticles. 07.04.2012
Extracellular glycosyl hydrolase activity of the Clostridium strains producing acetone, butanol, and ethanolO. V. Berezina, S. P. Sineoky, G. A. Velikodvorskaya, W. Schwarz, V. V. Zverlov ABSTRACT: Production of acetone, butanol, ethanol, acetic acid, and butyric acid by three strains of anaerobic bacteria, which we identified as Clostridium acetobutylicum, was studied. The yield of acetone and alcohols in 6% wheat flour medium amounted to 12.7–15 g/l with butanol constituting 51.0–55.6%. Activities of these strains towards xylan, β-glucan, carboxymethylcellulose, and crystalline and amorphous celluloses were studied. C. acetobutylicum 6, C. acetobutylicum 7, and C. acetobutylicum VKPM B-4786 produced larger amounts of acetone and alcohols and displayed higher cellulase and hemicellulase activities than the type strain C. acetobutylicum ATCC 824 in lab-scale butch cultures. It was demonstrated that starch in the medium could be partially substituted with plant biomass. Applied Biochemistry and Microbiology 04/2012; 44(1):42-47. · 0.56 Impact Factor 21.03.2012
Riboswitch control of Rho-dependent transcription terminationKerry Hollands, Sergey Proshkin, Svetlana Sklyarova, Vitaly Epshtein, Alexander Mironov, Evgeny Nudler, Eduardo A Groisman ABSTRACT: Riboswitches are RNA sensors that regulate gene expression upon binding specific metabolites or ions. Bacterial riboswitches control gene expression primarily by promoting intrinsic transcription termination or by inhibiting translation initiation. We now report a third general mechanism of riboswitch action: governing the ability of the RNA-dependent helicase Rho to terminate transcription. We establish that Rho promotes transcription termination in the Mg(2+)-sensing mgtA riboswitch from Salmonella enterica serovar Typhimurium and the flavin mononucleotide-sensing ribB riboswitch from Escherichia coli when the corresponding riboswitch ligands are present. The Rho-specific inhibitor bicyclomycin enabled transcription of the coding regions at these two loci in bacteria experiencing repressing concentrations of the riboswitch ligands in vivo. A mutation in the mgtA leader that favors the "high Mg(2+)" conformation of the riboswitch promoted Rho-dependent transcription termination in vivo and in vitro and enhanced the ability of the RNA to stimulate Rho's ATPase activity in vitro. These effects were overcome by mutations in a C-rich region of the mRNA that is alternately folded at high and low Mg(2+), suggesting a role for this region in regulating the activity of Rho. Our results reveal a potentially widespread mode of gene regulation whereby riboswitches dictate whether a protein effector can interact with the transcription machinery to prematurely terminate transcription. 07.03.2012
Metabolic engineering of Escherichia coli for 1-butanol biosynthesis through the inverted aerobic fatty acid β-oxidation pathway.Andrey Yu Gulevich; Alexandra Yu Skorokhodova; Alexey V Sukhozhenko; Rustem S Shakulov; Vladimir G Debabov Biotechnol. Lett. 34, 463 (2012) Research Institute for Genetics and Selection of Industrial Microorganisms, 1-st Dorozhniy Pr., 1, Moscow, Russia, 117545, gulevich@genetika.ru. The basic reactions of the clostridial 1-butanol biosynthesis pathway can be regarded to be the inverted reactions of the fatty acid β-oxidation pathway. A pathway for the biosynthesis of fuels and chemicals was recently engineered by combining enzymes from both aerobic and anaerobic fatty acid β-oxidation as well as enzymes from other metabolic pathways. In the current study, we demonstrate the inversion of the entire aerobic fatty acid β-oxidation cycle for 1-butanol biosynthesis. The constructed markerless and plasmidless Escherichia coli strain BOX-3 (MG1655 lacI ( Q ) attB-P( trc-ideal-4)-SD(φ10)-adhE(Glu568Lys) attB-P( trc-ideal-4)-SD(φ10)-atoB attB-P( trc-ideal-4)-SD(φ10)-fadB attB-P( trc-ideal-4)-SD(φ10)-fadE) synthesises 0.3-1 mg 1-butanol/l in the presence of the specific inducer. No 1-butanol production was detected in the absence of the inducer. 01.03.2012
Tissue regeneration in vivo within recombinant spidroin 1 scaffoldsMikhail M Moisenovich, Olga Pustovalova, Julia Shackelford, Tamara V Vasiljeva, Tatiana V Druzhinina, Yana A Kamenchuk, Vitaly V Guzeev, Olga S Sokolova, Vladimir G Bogush, Vladimir G Debabov, Mikhail P Kirpichnikov, Igor I Agapov ABSTRACT: One of the major tasks of tissue engineering is to produce tissue grafts for the replacement or regeneration of damaged tissue, and natural and recombinant silk-based polymer scaffolds are promising candidates for such grafts. Here, we compared two porous scaffolds made from different silk proteins, fibroin of Bombyx mori and a recombinant analog of Nephila clavipes spidroin 1 known as rS1/9, and their biocompatibility and degradation behavior in vitro and in vivo. The vascularization and intergrowth of the connective tissue, which was penetrated with nerve fibers, at 8 weeks after subcutaneous implantation in Balb/c mice was more profound using the rS1/9 scaffolds. Implantation of both scaffolds into bone defects in Wistar rats accelerated repair compared to controls with no implanted scaffold at 4 weeks. Based on the number of macrophages and multinuclear giant cells in the subcutaneous area and the number of osteoclasts in the bone, regeneration was determined to be more effective after the rS1/9 scaffolds were implanted. Microscopic examination of the morphology of the matrices revealed differences in their internal microstructures. In contrast to fibroin-based scaffolds, the walls of the rS1/9 scaffolds were visibly thicker and contained specific micropores. We suggest that the porous inner structure of the rS1/9 scaffolds provided a better micro-environment for the regenerating tissue, which makes the matrices derived from the recombinant rS1/9 protein favorable candidates for future in vivo applications. 08.02.2012
NotI Microarrays: Novel Epigenetic Markers for Early Detection and Prognosis of High Grade Serous Ovarian CancerVladimir Kashuba, Alexey A Dmitriev, George S Krasnov, Tatiana Pavlova, Ilya Ignatjev, Vasily V Gordiyuk, Anna V Gerashchenko, Eleonora A Braga, Surya P Yenamandra, Michael Lerman, Vera N Senchenko, Eugene Zabarovsky ABSTRACT: Chromosome 3-specific NotI microarray (NMA) containing 180 clones with 188 genes was used in the study to analyze 18 high grade serous ovarian cancer (HGSOC) samples and 7 benign ovarian tumors. We aimed to find novel methylation-dependent biomarkers for early detection and prognosis of HGSOC. Thirty five NotI markers showed frequency of methylation/deletion more or equal to 17%. To check the results of NMA hybridizations several samples for four genes (LRRC3B, THRB, ITGA9 and RBSP3 (CTDSPL)) were bisulfite sequenced and confirmed the results of NMA hybridization. A set of eight biomarkers: NKIRAS1/RPL15, THRB, RBPS3 (CTDSPL), IQSEC1, NBEAL2, ZIC4, LOC285205 and FOXP1, was identified as the most prominent set capable to detect both early and late stages of ovarian cancer. Sensitivity of this set is equal to (72 ± 11)% and specificity (94 ± 5)%. Early stages represented the most complicated cases for detection. To distinguish between Stages I + II and Stages III + IV of ovarian cancer the most perspective set of biomarkers would include LOC285205, CGGBP1, EPHB1 and NKIRAS1/RPL15. The sensitivity of the set is equal to (80 ± 13)% and the specificity is (88 ± 12)%. Using this technique we plan to validate this panel with new epithelial ovarian cancer samples and add markers from other chromosomes. 11.01.2012
Production of recombinant Rhizopus oryzae lipase by the yeast Yarrowia lipolytica results in increased enzymatic thermostability.Tigran V Yuzbashev, Evgeniya Y Yuzbasheva, Tatiana V Vibornaya, Tatiana I Sobolevskaya, Ivan A Laptev, Alexey V Gavrikov, Sergey P Sineoky ABSTRACT: The gene encoding Rhizopus oryzae lipase (ROL) was expressed in the non-conventional yeast Yarrowia lipolytica under the control of the strong inducible XPR2 gene promoter. The effects of three different preprosequence variants were examined: a preprosequence of the Y. lipolytica alkaline extracellular protease (AEP) encoded by XPR2, the native preprosequence of ROL, and a hybrid variant of the presequence of AEP and the prosequence of ROL. Lipase production was highest (7.6 U/mL) with the hybrid prepropeptide. The recombinant protein was purified by ion-exchange chromatography. The ROL included 28 amino acids of the C-terminal region of the prosequence, indicating that proteolytic cleavage occurred below the KR site through the activity of the Kex2-like endoprotease. The optimum temperature for recombinant lipase activity was between 30 and 40 °C, and the optimum pH was 7.5. The enzyme was shown not to be glycosylated. Furthermore, recombinant ROL exhibited greater thermostability than previously reported, with the enzyme retaining 64% of its hydrolytic activity after 30 min of incubation at 55 °C. Protein Expression and Purification 12/2011; 82(1):83-9. · 1.59 Impact Factor 12.12.2011
Efficient cell surface display of Lip2 lipase using C-domains of glycosylphosphatidylinositol-anchored cell wall proteins of Yarrowia lipolytica.Evgeniya Y Yuzbasheva, Tigran V Yuzbashev, Ivan A Laptev, Tatiana K Konstantinova, Sergey P Sineoky ABSTRACT: The cell surface display of enzymes is of great interest because of its simplified purification stage and the possibility for recycling in industrial processes. In this study, we have focused on the cell wall immobilization of Yarrowia lipolytica Lip2 protein--an enzyme that has a wide technological application. By genome analysis of Y. lipolytica in addition to already characterized Ylcwp1, we identified five putative open reading frames encoding glycosylphosphatidylinositol-anchored proteins. Lip2 translation fusion with the carboxyl termini of these proteins revealed that all proteins were capable of immobilizing lipase in active form on the cell surface. The highest level of cell-bound lipase activity was achieved using C-domains encoded by YlCWP1, YlCWP3 (YALI0D27214g) and YlCWP6 (YALI0F18282g) comprising 16,173 ± 1,800, 18,785 ± 1,130 and 17,700 ± 2,101 U/g dry cells, respectively. To the best of our knowledge, these results significantly exceed the highest cell-bound lipase activity previously reported for engineered Saccharomyces cerevisiae and Pichia pastoris strains. Furthermore, the lyophilized biomass retained the activity and was robust to collecting/resuspending procedures. Nevertheless, in most cases, a substantial amount of lipase activity was also found in the growth medium. Further work will be necessary to better understand the nature of this phenomenon. Applied Microbiology and Biotechnology 04/2011; 91(3):645-54. · 3.42 Impact Factor 21.03.2011
Is it possible to produce succinic acid at a low pH?Tigran V Yuzbashev, Evgeniya Y Yuzbasheva, Ivan A Laptev, Tatiana I Sobolevskaya, Tatiana V Vybornaya, Anna S Larina, Ilia T Gvilava, Svetlana V Antonova, Sergey P Sineoky [hide abstract] ABSTRACT: Bio-based succinate is still a matter of special emphasis in biotechnology and adjacent research areas. The vast majority of natural and engineered producers are bacterial strains that accumulate succinate under anaerobic conditions. Recently, we succeeded in obtaining an aerobic yeast strain capable of producing succinic acid at low pH. Herein, we discuss some difficulties and advantages of microbial pathways producing "succinic acid" rather than "succinate." It was concluded that the peculiar properties of the constructed yeast strain could be clarified in view of a distorted energy balance. There is evidence that in an acidic environment, the majority of the cellular energy available as ATP will be spent for proton and anion efflux. The decreased ATP:ADP ratio could essentially reduce the growth rate or even completely inhibit growth. In the same way, the preference of this elaborated strain for certain carbon sources could be explained in terms of energy balance. Nevertheless, the opportunity to exclude alkali and mineral acid waste from microbial succinate production seems environmentally friendly and cost-effective. 21.03.2011
Science, 23 April 2010, Volume 328: Cooperation Between Translating Ribosomes and RNA Polymerase in Transcription ElongationCooperation Between Translating Ribosomes and RNA Polymerase in Transcription Elongation Sergey Proshkin,1'2 A. Rachid Rahmouni,3 Alexander Mironov,z Evgeny Nudler1* Science, 23 April 2010, Volume 328, pp.504-508 During transcription of protein-coding genes, bacterial RNA polymerase (RNAP) is closely followed by a ribosome that translates the newly synthesized transcript. Our in vivo measurements show that the overall elongation rate of transcription is tightly controlled by the rate of translation. Acceleration and deceleration of a ribosome result in corresponding changes in the speed of RNAP. Moreover, we found an inverse correlation between the number of rare codons in a gene, which delay ribosome progression, and the rate of transcription. The stimulating effect of a ribosome on RNAP is achieved by preventing its spontaneous backtracking, which enhances the pace and also facilitates readthrough of roadblocks in vivo. Such a cooperative mechanism ensures that the transcriptional yield is always adjusted to translational needs at different genes and under various growth conditions.
03.10.2010
Isolation ofanewbutanol-producing Clostridium strain: Highlevel of hemicellulosicactivityandstructureofsolventogenesisgenes of anew Clostridium saccharobutylicum isolateOksana V.Berezinaa,, AgnieszkaBrandtb, SergeyYarotskya, Wolfgang H.Schwarzb, VladimirV.Zverlovb, cState ResearchInstituteofGeneticsandSelectionofIndustrialMicroorganisms,1stDorojniypr.1,117545Moscow, Russian Federation bDepartment ofMicrobiology,TechnischeUniversita¨t Mu¨nchen, AmHochanger4,85350Freising,Germany cInstitute ofMolecularGenetics,RussianAcademyofScience,KurchatovSq.2,123182Moscow,RussianFederation
Abstract New isolates of solventogenic bacteria exhibitedhighhemicellulolyticactivity.Theyproducedbutanoland acetone withhighselectivityforbutanol(about80%ofbutanolfromthetotalsolventyield).Their16SrDNA sequencewas99%identicaltothatof Clostridiumsaccharobutylicum. Thegenesresponsibleforthelaststeps of solventogenesisandencodingcrotonase,butyryl-CoAdehydrogenase,electron-transportproteinsubunitsAandB, 3-hydroxybutyryl-oAdehydrogenase,alcoholdehydrogenase,CoA-transferase(subunitsAandB),acetoacetate decarboxylase,andaldehydedehydrogenasewereidentifiedinthenew C. saccharobutylicum strainOx29andcloned into Escherichiacoli. The genesforcrotonase,butyryl-CoAdehydrogenase,electron-transportproteinsubunitsA and B,and3-hydroxybutyryl-CoAdehydrogenasecomposedthe bcs-operon.Amonocistronicoperoncontaining the alcoholdehydrogenase gene was located downstream of the bcs-operon.Genesforaldehydedehydrogenase, CoA-transferase(subunitsAandB),andacetoacetatedecarboxylasecomposedthe sol-operon. The gene sequences and thegeneorderwithinthe sol- and bcs-operons of C. saccharobutylicum Ox29 were most similar to those of Clostridiumbeijerinckii. The activity of some of the bcs-operon genes,expressed in heterologous E. coli, was determined. 12.08.2010
Production of succinic acid at low pH by a recombinant strain of the aerobic yeast Yarrowia lipolytica.Tigran V Yuzbashev, Evgeniya Y Yuzbasheva, Tatiana I Sobolevskaya, Ivan A Laptev, Tatiana V Vybornaya, Anna S Larina, Kazuhiko Matsui, Keita Fukui, Sergey P Sineoky [hide abstract] ABSTRACT: Biotechnological production of weak organic acids such as succinic acid is most economically advantageous when carried out at low pH. Among naturally occurring microorganisms, several bacterial strains are known to produce considerable amounts of succinic acid under anaerobic conditions but they are inefficient in performing the low-pH fermentation due to their physiological properties. We have proposed therefore a new strategy for construction of an aerobic eukaryotic producer on the basis of the yeast Yarrowia lipolytica with a deletion in the gene coding one of succinate dehydrogenase subunits. Firstly, an original in vitro mutagenesis-based approach was proposed to construct strains with Ts mutations in the Y. lipolytica SDH1 gene. These mutants were used to optimize the composition of the media for selection of transformants with the deletion in the Y. lipolytica SDH2 gene. Surprisingly, the defects of each succinate dehydrogenase subunit prevented the growth on glucose but the mutant strains grew on glycerol and produced succinate in the presence of the buffering agent CaCO(3). Subsequent selection of the strain with deleted SDH2 gene for increased viability allowed us to obtain a strain capable of accumulating succinate at the level of more than 45 g L(-1) in shaking flasks with buffering and more than 17 g L(-1) without buffering. The possible effect of the mutations on the utilization of different substrates and perspectives of constructing an industrial producer is discussed. Biotechnology and Bioengineering 11/2010; 107(4):673-82. · 3.95 Impact Factor 21.03.2010
A novel model system for design of biomaterials based on recombinant analogs of spider silk proteinsBogush VG, Sokolova OS, Davydova LI, Klinov DV, Sidoruk KV, Esipova NG, Neretina TV, Orchanskyi IA, Makeev VY, Tumanyan VG, Shaitan KV, Debabov VG, Kirpichnikov MP. 05.01.2008
Screening, Characterization and Application of Cyanide-resistant Nitrile HydratasesT. Gerasimova, A. Novikov, S. Osswald, A. Yanenko
05.08.2004
A Cluster of Thermotoga neapolitana Genes Involved in the Degradation of Starch and Maltodextrins: the Molecular Structure of the LocusO. V. Berezina, N. A. Lunina, V. V. Zverlov, D. G. Naumoff, W. Liebl, G. A. Velikodvorskaya ABSTRACT: A 5451-bp genome fragment of the hyperthermophilic anaerobic eubacterium Thermotoga neapolitana has been cloned and sequenced. The fragment contains one truncated and three complete open reading frames highly homologous to the starch/maltodextrin utilization gene cluster from T. maritima whose genome sequence is known. The incomplete product of the first frame is highly homologous to MalG, the Escherichia coli protein of starch and maltodextrin transport. The product of the second frame, AglB, is highly homologous to cyclomaltodextrinase with the -glucosidase activity TMG belonging to family 13 of glycosyl hydrolases (GH13). The product of the third frame, AglA, is homologous to the T. maritima cofactor-dependent -glucosidase from the GH4 family. The two enzymes form a separate branch on the phylogenetic tree of the family. The AglA and AglB proteins supplement each other in substrate specificity and can ensure complete hydrolysis to glucose of cyclic and linear maltodextrins, the intermediate products of starch degradation. The product of the fourth reading frame has sequence similarity with the riboflavin-specific deaminase RibD from T. maritima. The homologous locus of this bacterium, between the aglA and ribD genes, has five open reading frames missing in T. neapolitana. The nucleotide sequences of two frames are homologous to transposase genes. The deletion size is 2.9 kb. Molecular Biology 08/2003; 37(5):678-685 07.08.2003
DcrA and dcrB Escherichia coli genes can control DNA injection by phages specific for BtuB and FhuA receptors.Valery V Samsonov, Victor V Samsonov, Sergey P Sineoky [hide abstract] ABSTRACT: We had previously shown that the Escherichia coli proteins DcrA (SdaC) and DcrB, located, respectively, in the inner membrane and periplasm, are involved in the early development of virulent bacteriophage C1, which recognises BtuB as an outer membrane receptor. In the present work it is demonstrated that the DcrA and DcrB proteins, coordinately with another outer membrane receptor protein, FhuA, are also involved in an early stage of development of a newly isolated virulent phage, C6. In both cases, DcrA and DcrB probably are required in the second stage of phage adsorption-the DNA injection process. This means that DcrA and DcrB proteins can participate in phage DNA transport pathways in cooperation with different outer membrane receptors, including FhuA and BtuB. The increased sensitivity of bacterial cells to SDS following C1 and C6 adsorption suggests that adsorption by these phages triggers the opening of diffusion channels through the outer membrane. Our results also indirectly demonstrate that the DcrA and DcrB proteins participate in the opening or formation of these channels. 21.03.2003
The Threonine StoryVladimir G. DebabovAdvances in Biochemical Engineering/
Biotechnology,Vol. 79
Managing Editor: T. Scheper
© Springer-Verlag Berlin Heidelberg 2003
Advances in biochemical engineering/biotechnology 79:113-36, 2003 05.01.2003
Cloning and expression of the lux-operon of Photorhabdus luminescens, strain Zm1: nucleotide sequence of luxAB genes and basic properties of luciferaseI V Manukhov, S M Rastorguev, G E Eroshnikov, A P Zarubina, G B Zavil'gel'skiĭ ABSTRACT: A chromosomal fragment of bacteria Photorhabdus luminescence Zm1, which contains the lux operon, was cloned into the vector pUC18. The hybrid clone containing plasmid pXen7 with the EcoRI fragment approximately 7-kb was shown to manifest a high level of bioluminescence. By subcloning and restriction analysis of the EcoRI fragment, the location of luxCDABE genes relative to restriction sites was determined. The nucleotide sequence of the DNA fragment containing the luxA and luxB genes encoding alpha- and beta-subunits of luciferase was determined. A comparison with the nucleotide sequences of luxAB genes in Hm and Hw strains of Ph. luminescence revealed 94.5 and 89.7% homology, respectively. The enterobacterial repetitive intergenic sequence (ERIC) of 126 bp typical for Hw strains was identified in the spacer between the luxD and luxA genes. The lux operon of Zm1 is assumed to emerge through recombination between Hm and Hw strains. Luciferase of Ph. luminescence was shown to possess a high thermal stability: its activity decreased by a factor of 10 at 44 degrees C for 30 min, whereas luciferases of marine bacteria Vibrio fischeri and Vibrio harveyi were inactivated by one order of magnitude at 44 degrees C for 1 and 6 min, respectively. The lux genes of Ph. luminescence are suggested for use in gene engineering and biotechnology. Genetika 04/2000; 36(3):322-30. 16.03.2000
Nitrile Hydratase of Rhodococcus. Optimization of Synthesis in Cells and Industrial Applications for Acrylamide ProductionTATYANA E. LEONOVA (POGORELOVA), OLGA B. ASTAUROVA, LUDMILA E. RYABCHENKO, AND ALEXANDER S. YANENKO*05.01.2000
RecA-independent pathways of lambdoid prophage induction in Escherichia coli.D V Rozanov, R D'Ari, S P Sineoky ABSTRACT: Two Escherichia coli genes, expressed from multicopy plasmids, are shown to cause partial induction of prophage lambda in recA mutant lysogens. One is rcsA, which specifies a positive transcriptional regulator of the cps genes, which are involved in capsular polysaccharide synthesis. The other is dsrA, which specifies an 85-nucleotide RNA that relieves repression of the rcsA gene by histone-like protein H-NS. Genetic contexts known to increase Cps expression also cause RecA-independent lambda induction: the rcsC137 mutation, which causes constitutive Cps expression, and the lon and rcsA3 mutations, which stabilize RcsA. Lambdoid phages 21, phi80, and 434 are also induced by RcsA and DsrA overexpression in recA lysogens. Excess lambda cI repressor specifically blocks lambda induction, suggesting that induction involves repressor inactivation rather than repressor bypass. RcsA-mediated induction requires RcsB, the known effector of the cps operon, whereas DsrA-mediated induction is RcsB independent in stationary phase, pointing to the existence of yet another RecA-independent pathway of prophage induction. Journal of Bacteriology 01/1999; 180(23):6306-15. · 3.83 Impact Factor 21.03.1999
Article: Genetic control of the resistance to phage C1 of Escherichia coli K-12.N A Likhacheva, V V Samsonov, S P Sineoky ABSTRACT: Escherichia coli K-12 lytic phage C1 was earlier isolated in our laboratory. Its adsorption is controlled by at least three bacterial genes: dcrA, dcrB, and btuB. Our results provide evidence that the dcrA gene located at 60 min on the E. coli genetic map is identical to the sdaC gene. This gene product is an inner membrane protein recently identified as a putative specific serine transporter. The dcrB gene, located at 76.5 min, encodes a 20-kDa processed periplasmic protein, as determined by maxicell analysis, and corresponds to a recently determined open reading frame with a previously unknown function. The btuB gene product is known to be an outer membrane receptor protein responsible for adsorption of BF23 phage and vitamin B12 uptake. According to our data the DcrA and DcrB proteins are not involved in these processes. However, the DcrA protein probably participates in some cell division steps. Journal of Bacteriology 10/1996; 178(17):5309-15. · 3.83 Impact Factor 21.03.1996
Single amino acid substitutions in the cAMP receptor protein specifically abolish regulation by the CytR repressor in Escherichia coli.L Søgaard-Andersen, A S Mironov, H Pedersen, V V Sukhodelets, P Valentin-Hansen ABSTRACT: Promoters in Escherichia coli that are negatively regulated by the CytR repressor are also activated by the cAMP receptor protein (CRP) complexed to cAMP; as a characteristic, these promoters encode tandem binding sites for cAMP-CRP. In one such promoter, deoP2, CytR binds to the region between the tandem CRP binding sites with a relatively low affinity; in the presence of cAMP-CRP, however, the repressor and activator bind cooperatively to the DNA. Here we have investigated this cooperativity by isolating mutants of the CRP protein that abolish CytR regulation without exhibiting a concomitant loss in their ability to activate transcription. Four different, single amino acid substitutions in CRP give rise to this phenotype. These amino acids lie in close proximity on the surface of the CRP tertiary structure in a portion of the protein that is not in contact with the DNA. In vitro analyses of one of the CRP mutants show that it interacts with the DNA in a manner indistinguishable from wild-type CRP, whereas its interaction with CytR is perturbed. These results strongly indicate that cooperative DNA binding of CytR and cAMP-CRP is achieved through protein-protein interactions. Proceedings of the National Academy of Sciences 07/1991; 88(11):4921-5. 13.03.1991
|